Acoustic detection of a mutation-specific Ligase Chain Reaction based on liposome amplification
Authors: Nikoletta Naoumi, Monica Araya-Farias, Maria Megariti, Lucile Alexandre, George Papadakis, Stephanie Descroix, and Electra Gizeli
Journal: Analyst
Abstract
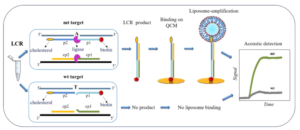
Single nucleotide variants (SNVs) play a crucial role in understanding genetic diseases, cancer development, and personalized medicine. However, existing ligase-based amplification and detection techniques, such as Rolling Circle Amplification and Ligase Detection Reaction, suffer from low efficiency and difficulties in product detection. To address these limitations, we propose a novel approach that combines Ligase Chain Reaction (LCR) with acoustic detection using highly dissipative liposomes. In our study, we are using LCR combined with biotin- and cholesterol-tagged primers to produce amplicons also modified at each end with a biotin and cholesterol molecule. We then apply the LCR mix without any purification directly on a neutravidin modified QCM device Au-surface, where the produced amplicons can bind specifically through the biotin end. To improve sensitivity, we finally introduce liposomes as signal enhancers. For demonstration, we used the detection of the BRAF V600E point mutation versus the wild-type allele, achieving an impressive detection limit of 220 aM of the mutant target in the presence of the same amount of the wild type. Finally, we combined the assay with a microfluidic fluidized bed DNA extraction technology, offering the potential for semi-automated detection of SNVs in patients’ crude samples. Overall, our LCR/acoustic method outperforms other LCR-based approaches and surface ligation biosensing techniques in terms of detection efficiency and time. It effectively overcomes challenges related to DNA detection, making it applicable in diverse fields, including genetic disease and pathogen detection.